Research
Screening for Primordial RNA–Peptide Interactions Using High-Density Peptide Arrays
|
Highlights:
See for details: MDPI article The combinatorial fusion cascade: MDPI article
Figure: High-throughput screening of RNA–peptide interactions
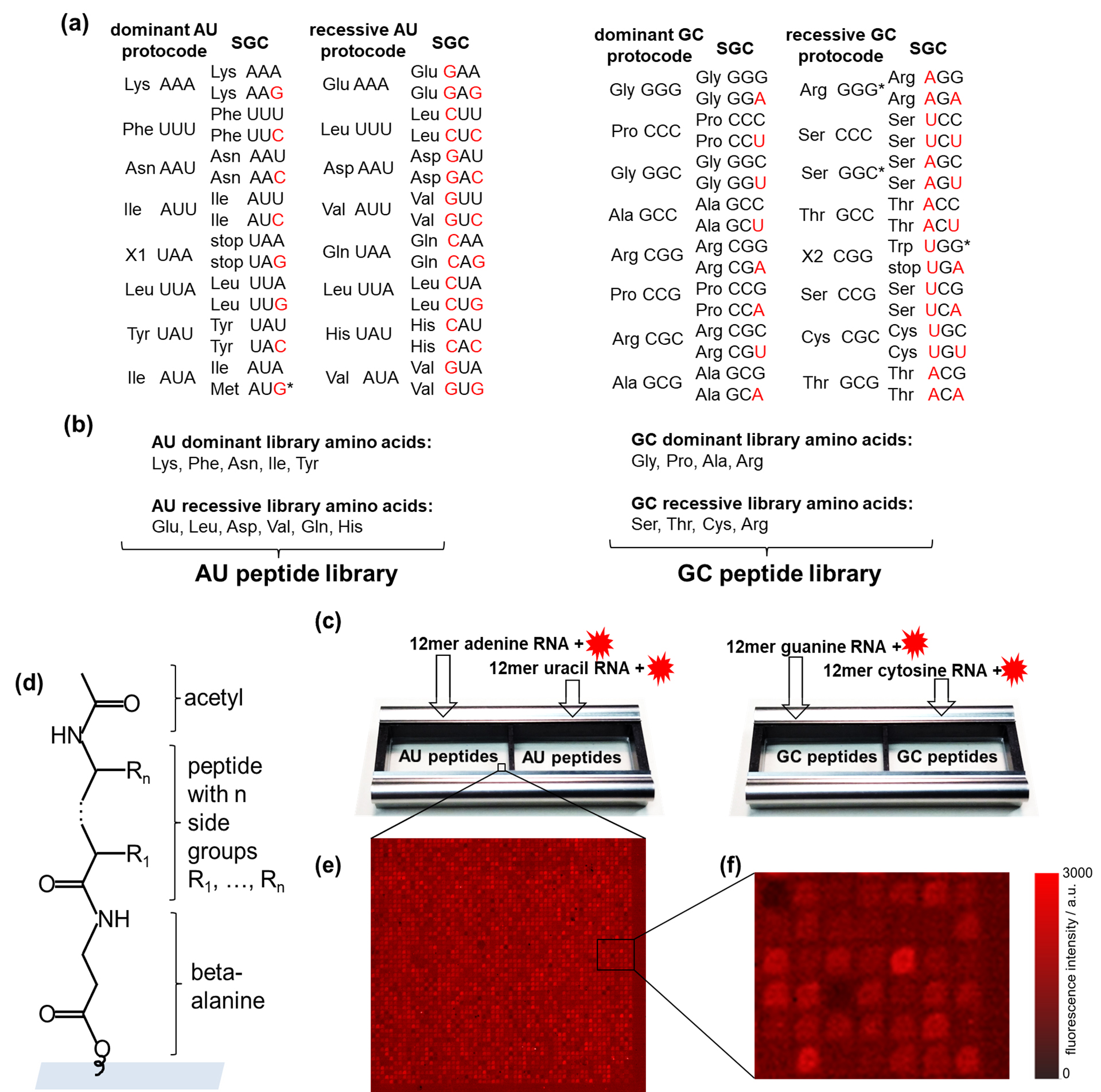
(a) AU- and GC-protocodes and their transformation to the standard genetic code (SGC) after the combinatorial fusion. The codons of the SGC were obtained according to the Watson–Crick mutations A↔G or U↔C in the third position for the dominant protocodes. Watson–Crick mutations for recessive protocodes occurred in the first or the first and third positions. The red letters illustrate these transformations. (b) The AU-protocode library consists of two sublibraries: all combinatorial combinations of the amino acids of the dominant protocode (Lys, Phe, Asn, Ile, and Tyr) and all combinatorial combinations of the amino acids of the recessive protocode (Glu, Leu, Asp, Val, Gln, and His). The GC-protocode library consists of two sublibraries: all combinatorial combinations of the amino acids of the dominant protocode (Gly, Pro, Ala, and Arg) and all combinatorial combinations of the amino acids of the recessive protocode (Ser, Thr, Cys, and Arg). (c) Location of the peptide libraries in the incubation trays and their incubation with fluorescently labeled 12-mer single-stranded RNA homo oligomers of adenine, guanine, uracil, and cytosine. (d) Peptide structure in the spot: beta-alanine as linker, acetylated N-terminus. (e,f) Fragments of fluorescence images of peptide chips after incubation with fluorescently labeled RNAs. The pitch size was 60 m. |
|
Resemblance-Ranking Peptide Library to Screen for Binders to Antibodies on a Peptidomic Scale
|
Highlights:
See for details: MDPI article
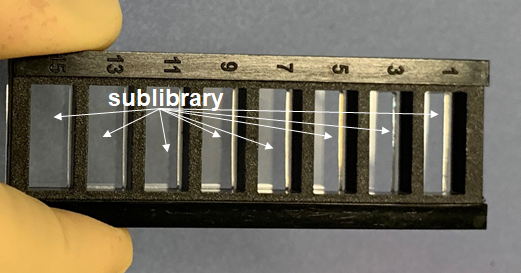
Figure: Eight wells with the same sub-library of 6000 different peptides for determining dissociation constants by incubating antibodies at different concentrations. |
Serological Number for Characterization of Circulating Antibodies
|
Highlights:
See for details: MDPI article
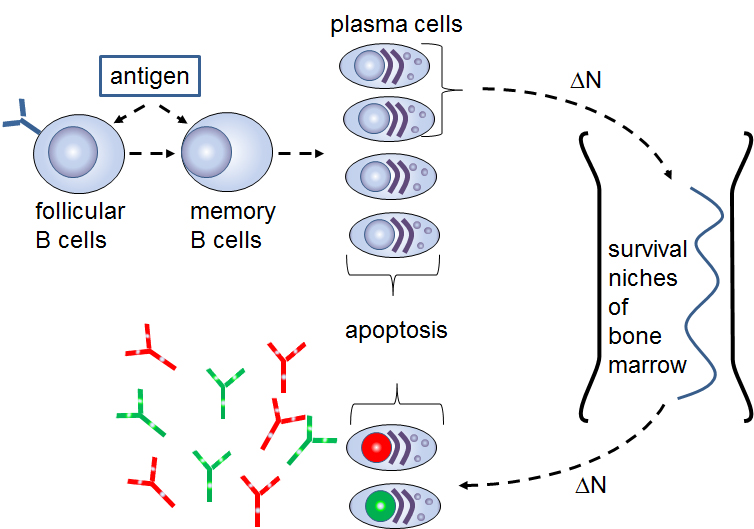
Figure: Schematic of the long-term humoral memory via elimination of the long-lived plasma cells |
Selective Peptide Binders to the Perfluorinated Sulfonic Acid Ionomer Nafion
|
Highlights:
See for details: Wiley article
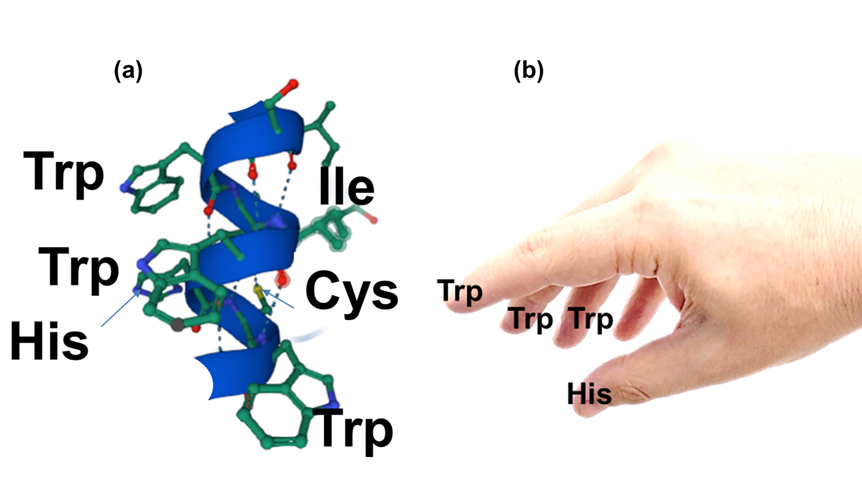
Figure: Structure of the peptide WIWHCW with a selective binding to Nafion. a) Helix structure of the peptide WIWHCW. b) Schematic presentation of the selective binding geometry as a gripping hand with Trp (W) and His (H) side chains as fingers. |
|